A recent paper published by the Answers Research Journal, the research publication of Answers in Genesis, reported a comparison of human and chimpanzee genomes and found that they have, on average, a DNA similarity of only 70%. This is a very striking number since the usual numbers you hear thrown about as representing the similarity of the human and chimpanzee genome are usually 98.6%, 98%, 96%, 94% and even 90%. The variation in these values is partly due to larger data sets becoming available for comparison but is mostly due to different assumptions used to calculate the percentages. For example, the higher similarities are derived by examining only DNA sequences that correspond to actual pieces of genetic code (ie. the genes in the genome). The estimates of lower similarity reflect comparisons that include vast stretches of DNA that don’t include coding regions or are the spacers between genes. Now comes word from a paper published by Answers in Genesis that the average similarity of the DNA sequence is actually only 70%. What is up with that? Is this as significant a finding as it sounds? (see footnote)
First, let me say I am really not interested in entering into a protracted discussion about human origins. I not saying that humans aren’t a unique creation. Rather I am commenting on this paper because I think that it promotes a very poor scientific argument for the uniqueness of man and thus is one that Creationists should stop using. Tompkin and others at ICR and AIG have striven for many years to demonstrate that chimpanzees and humans could not be related because they are far too different, both genetically and morphologically, for anyone to reasonably believe they shared a common ancestor. They have attempted to build a genetic case against evolution and promote these differences as proofs that give the lay Christian confidence that there is no connection between humans and chimpanzees.
 In this brief article I will argue that rather than providing strong case against primate-human ancestry, Tomkins and others actually undermine their own case for the uniqueness of man by their inconsistent approach to applying genetic distance as a principle of species (or “kinds”) delineation. Let’s start by looking at the conclusion of Tomkin’s article from Feb. 20, 2013: “Comprehensive Analysis of Chimpanzee and Human Chromosomes Reveals Average DNA Similarity of 70%.”
In this brief article I will argue that rather than providing strong case against primate-human ancestry, Tomkins and others actually undermine their own case for the uniqueness of man by their inconsistent approach to applying genetic distance as a principle of species (or “kinds”) delineation. Let’s start by looking at the conclusion of Tomkin’s article from Feb. 20, 2013: “Comprehensive Analysis of Chimpanzee and Human Chromosomes Reveals Average DNA Similarity of 70%.”
“Genome-wide, only 70% of the chimpanzee DNA was similar to human under the most optimal sequence-slice conditions. While, chimpanzees and humans share many localized protein-coding regions of high similarity, the overall extreme discontinuity between the two genomes defies evolutionary timescales and dogmatic presuppositions about a common ancestor.”
Tomkins makes it abundantly clear that he believes that such extreme genetic dissimilarity between humans and chimps should make it ridiculous for anyone to think that they humans and chimpanzees could have a common ancestor. How does he come to this 70% number when other geneticists derive much higher values. First, Tomkins states quotes a “geneticist” named Buggs that apparently had already calculated a 76% similarity between the genomes. The implication seems to be that scientists already knew that there was a big difference but have been ignoring it. However, I tried to follow the link in the references to this Buggs quote but the link (only 7 days after publication!) is already dead or was printed in error. (see comment below for reader supplied links). Regarding how Tomkins derives the 70% value, I really am not certain because the methods described in the paper do not allow me to fully ascertain how the analyses was done. However, Tomkins clearly is comparing all parts of the genome and depending on how they align is counting not only differences in the sequences, but gaps where there are insertions and deletions between the genomes, repeat units that differ between the genomes and inversions (pieces of the genome that are flipped around thus making the genes appear in different arrangements between genomes). I really don’t need to get bogged down into describing all these part of a genome structure here because the methods he used are really not that important. Why? Because the method he used doesn’t really matter to me. I would not deny the he derived this 70% value with the algorithm he used. In fact, does the 70% surprise me? Not really. There are huge tracks (hundreds of millions of base pairs) of the human genome that are repeated sections and pseudogenes (broken, unused genes) and those regions have very low sequence similarity to other animals. With Tomkins way of counting differences I would expect there to be huge numbers of differences. In fact, you and I may only be 80% similar in some regions of our DNA. So what’s the big deal then?
70%, 90%, 98% – what’s in a number?
Well, really percentages don’t matter that much. Who cares if something is 90% similar with a given algorithm or it is 98% similar. What matters much much more is how those numbers compares to other similar pairwise comparisons. In other words, you need to do an apples-to-apples, or oranges-to-oranges if you prefer, analyses to actually be able to compare something meaningfully. So let’s take Tomkins 70% number. That number makes it sound like there is an extreme difference in the DNA sequences of humans and chimps especially when compared to the 96-98% number that has been repeated so much in the literature. But does this mean that humans are so much more different from chimpanzees than scientists have claimed. Honestly, when I began reading this paper the first thing I thought was, this 70% number is completely meaningless unless Tomkins provides me with a context for that number. When I reached the end I found that he provided NO context at all. He continued to talk only of human-chimp comparisons and never once compared his values with the genetic similarities, based on the same methods, of other animals or even among other humans. Without doing so he has done little to nothing to make a case that “the overall extreme discontinuity between the two genomes defies evolutionary timescales.” Why? Because he gave me no idea if 70% really is a lot of difference or not? He is hoping the reader will be astounded by the 70% and never ask, what should be obvious, if 70% really is a lot of difference or does his method of accounting result in all species looking very different?
Tomkins own analysis provides evidence that conflict with his and other creationists’ conclusions about common ancestry!
I asserted above that Tomkins and others are actually undercutting their own beliefs in the genetic uniqueness of man. Where do I get that from? It goes back to the creationist concept of baraminology (See my review of baraminology – Some thoughts on Baraminology). Creationists, such as Tomkins, today are very likely to promote the view of super-speed evolution after the Food. For example, at the AIG Creation Museum, there is a graphic that shows all canine species alive today descending from a pair of dogs on the Ark. Similarly, creationists studying baramins (the original created kinds) have generally concluded that all cats came from a common ancestor. I am writing more about the ostrich and it appears that most creationists think that the ostrich could fly and that the 20 species of extinct Moa’s on New Zealand were once a single Moa that flew to New Zealand and then evolved there. In a nutshell, creationists believe that 100s of thousands of species on this earth are descendants of common ancestors that were found in pairs on Noah’s ark.
Coming back to the 70% value, what does this have to do with baraminology? Tomkins would like his 70% number to sound like such a difference that it would impossible to believe that anything that dissimilar could have a common ancestor. If he believes this is true he could do some very simple analyses to test his hypothesis. His hypothesis could be used to make many predictions but he fails to make any. He could propose tests to see if his 70% number is really a significant value. The biggest test he could propose would be to compare other genomes of other organisms that he believed did descend from a common ancestor. His model would predict that they would show much greater (ie >70%) similarity in their DNA sequences using the same algorithm he used.
When I read Tomkins paper I thought to myself, if chimps and humans are only 70% similar then how similar is a modern human to a Neanderthal or to the Denisovan sequence? For that matter, how similar is one modern human to another. Tomkins provides us with no clue here even though he could have done the analysis. I think this is because he doesn’t want to provide a distraction from his glaring 70% number. I know that those that say that humans and chimps are 96% similar would also find that all modern humans are at least 99% similar to each other and 98% similar to Denisovans and a bit more similar to Neanderthals. Why doesn’t Tomkins apply his same algorithm to other humans sequences and Neanderthals? Since he didn’t it is hard to compare his numbers to other humans and to compare species in other baramins (kinds). What if Tomkins used his algorithm to look at the human and Denisovan genomes and found that they were 90% or even 85% similar. Would he then be able to confidently say that Denisovan’s really were the same as modern humans? What if he compared individual humans and found that that you and I are only 95% similar? Would the 70% value seem as dramatic if modern humans and Neanderthal’s where only 85% similar?
Now here comes the big question: What if Tomkins used the same assumptions for calculating DNA sequence similarity among dog species or cat species and found that they were more different than humans and chimps? What if some of them are only 60% similar? If they are more dissimilar can Tomkins really erect a genetic similarity argument that says that canines all evolved from a common ancestor in 4000 years and are more genetically dissimilar than chimps and humans which he emphatically believes didn’t have a common ancestor? I wonder if Tompkins has ever really thought about the implications of his own data. He has the ability to test his model but makes no mention of what he would expect to find in other species? Why? I get the feeling that Tomkins and others know they are playing with a double standard. They are promoting rapid evolution, including radical genetic change over a short period of time, from common ancestors from one side of their mouth while at the same time proclaiming that genetic dissimilarity precludes common ancestry in other groups from the other side of their mouth.

Of Cats, Dogs and Primates….
I can’t use Tomkins calculations to do my own analyses but based on my own personal experience having worked with a LOT of DNA sequences and done thousands of comparisons of DNA sequences across many groups of organisms I think I have a pretty good idea of what the overall differences between various animals groups would be. So I will make a few predictions right now and then I will collect some data to see if they confirm my suspicions. I will report my findings in my next post on this topic.
I predict that I will be able to find multiple organisms that are demonstrate greater genetic differences than humans and chimpanzees but that
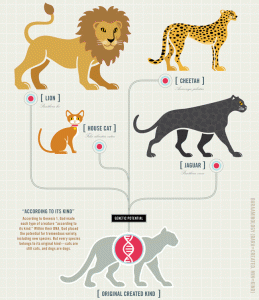
Tomkins and other creationists are likely to believe share a common ancestor. The species that I will target for my analyses will be those in the canine and feline families because creation scientists seem to believe that these two groups each were founded by a single pair of ancestors.
I don’t’ have the resources, nor is there enough data in some cases, for me to compare entire nuclear genomes of these organisms but it is relatively easy for anyone to access entire mitochondrial genomes from thousands of animals and determine their genetic similarity. All animal cells have a separate genome in their mitochondria which is much smaller than their nuclear genome. In humans this genome is 16,569 bases (As,Ts,Cs and Gs). It is similar size in all mammals so it is fairly easy to compare and 10s of thousands of entire genomes have been sequenced. For organisms that aren’t too different from one another differences in their mitochondrial genomes represent a fairly good proxy for the scale of differences I would expect to find in their nuclear genomes (see footnote). Here is something that I do know (based on data from Wikipedia) right now about the mitochondrial DNA of primates:
1) Chimpanzees mitochondrial genomes differ from the average human by 1462 base pairs out of 16,569 total bases in humans. That is an 8.9% difference (or 91.1% similarity). Notice that Tomkins found a 70% similarity for the nuclear genome but the mtDNA genome is easy to compare and so this number is pretty solid and agreed upon by everyone.
2) The average human mitochondrial genome differs from the Denisovan (fossil human) by 385 bp (2.3%)
3) The average human mitochondrial genome differs from the Neanderthal (fossil human) by 202 bp (1.2%)
4) I am not sure how different individual humans are but I’m going to guess that any two individual humans alive today differ by 0 to 75 bp. (ie. less than 0.5%)
I am sure Tomkins would point to the data above and feel confident that it shows that humans and chimps are genetically very different. I would agree, but if he is right that these genetic distance values tell us that humans are not related by ancestry to chimps then if we extend his assumptions to other animals we should expect to see that similar organisms will show much less than an 8.9% difference between them. But here is why I think that Tomkins is in big trouble if he were to actually attempt to do an apples-to-apples comparison of difference species using his methods and assumptions.
Here are some predictions of what I am going to find when I compare mitochondrial genomes within a number of animals groups:
1) Foxes and wolves will be at least as different as chimpanzees and humans and possibly have more differences in the mitochondrial genomes.
2) Domestic dogs will have less variation in their mitochondrial genomes than do humans
3) Domestic cats and/or leopards and cheetahs will be more different than humans and chimpanzees
4) Tigers and lions, which can interbreed, maybe as genetically different as humans and chimpanzees
5) I think that many species of bears may have more differences in their mitochondrial genomes than do chimpanzees and humans. I think the Panda which creationists will say is a bear “kind” will be more different than chimpanzees and humans.
6) Apes, orangutans and chimpanzees which are all placed in the same “kind” by creationist will be more different than one another than chimpanzees will be from humans.
If I am correct in my predictions then it should be apparent that Tomkins and other need to seriously think about how they wish to equate genetic similarity with species boundaries. I’ll start crunching some numbers and come back with a report soon.
Notes: I have done the analysis and and written about it: Of Kinds and Common Ancestors: Comparing Mitochondrial Genomes
Footnotes:
DNA sequence similarity: Before someone sends me a comment about DNA similarity not being the only factor in determining genetic relatedness let me assure you that I am aware that two sequences that are very similar can either have the same function or completely different functions. Sometimes just a single change can make a huge difference but in other genes the sequences could be 20% different and the gene would still perform its function in the same way. So gross estimates of genetic similarity are not foolproof ways of assessing the differences between organisms. Since the Tomkins paper is making a case that sequencing differences are too great for there to be common ancestry that is the specific issue I’m interested in exploring right now.
mtDNA: I am fully aware that mtDNA sequences “evolve” at a faster rater than do sequences of nuclear genes.However, compared to the Tomkin’s estimations mtDNA sequences are actually more similar to each other than the nuclear genome. Differences in mtDNA sequences are a rough approximation of how different the whole nuclear genome will be. For example if there is a 9% difference in mtDNA sequence then there is typically going to be about a 2% difference in the coding region of the nuclear genes but if there was 18% difference in the mtDNA there would be bout a 4% difference in the nuclear genome. At much higher rates of differences the differences don’t equate as well but there isn’t enough room here to explain why this is other than to say it has to do with homoplasy for those that know the term.
Hi, you can read Dr. Bugg’s two articles here: http://web.archive.org/web/20090302001800/http://www.refdag.nl/artikel/1366432/Chimpanzee.html and http://web.archive.org/web/20081211153345/http://www.refdag.nl/artikel/1378077/70+Chimp.html. More about Buggs here: http://www.richardbuggs.com/
LikeLike
Thanks for those links. They are very helpful. The first provides a better description of where that 70% is coming from. What I found interesting is that he does note that for the sequence (2.4 billion base pairs) that do align between chimp and human there is a 1.23% difference. This is where the 98% value is coming from. So all these other differences are in addition to the actual individual base pair changes. If I understand right, both Buggs and Tomkins are looking at a pieces of sequence and if both chimp and human share 1 million base pairs but there are an extra 1 million base pairs in humans that have no equivalent sequence or just can’t be aligned even if there is some extra sequence in both then they would call this 50% overall similarity. Not all that surprising really but using this form of accounting for similarity will lead to some weird results. The question really is whether overall similarity of sequence means anything. My Y chromosome has many sections that have very low similarity to other males but in some regions that code for genes the sequences are very very similar. Its what genes are present, what they do and how they coordinate with each other in development that made the biggest biological difference in how we view differences between species. Thanks again for those links.
LikeLike
Hi, I know this is an old thread but I love coming back to read this excellent post. I do have a question, though. In your comment you said that:
If I understand right, both Buggs and Tomkins are looking at a pieces of sequence and if both chimp and human share 1 million base pairs but there are an extra 1 million base pairs in humans that have no equivalent sequence or just can’t be aligned even if there is some extra sequence in both then they would call this 50% overall similarity. Not all that surprising really but using this form of accounting for similarity will lead to some weird results.
Could you clarify what these weird results would be? I was wondering if it had anything to do with the C-paradox where even some closely related species have genome sizes that vary wildly for example, from the blog Genomicron http://www.genomicron.evolverzone.com/2007/04/onion-test/
From the above link, it would seem that by the method of similarity used by Buggs and Tomkins, the onions A. altyncolicum and A. ursinum would have an overall similarity of << 70%.
LikeLike
I’ve had a chance to talk to a few other people that have really tried to understand Tomkins paper and recreate his analysis. They were as frustrated as I was in determining what he actually did. But I think you have the gist of it there. Large insertions or deleations seem to be treated not as single losses or gains but rather a 100bp insertion is treated as 100 differences. In an evolutionary model I suppose it technically could be 100 individual insertions of 1 base each but the most parsimonious explanation would be a one time 100 bp insertion/duplication event which is usually treated as a single character difference. You have seen how this can become quite absurd when two species that do have very different genomes sizes are compared. Massive duplication via transposable elements could create huge numbers of insertions or deletions relative to a closely related species. Tomkins analysis would suggest a very low overall similarity. I’ve suggested to contacts that know Tomkins that he compare two different modern human genomes with the exact same criteria and report his results. Better yet compare a dog and fox (not sure there is a whole genome yet of a fox but I am sure there will be soon) which are said to have been one “kind” on the ark and see what the difference is there. I expect it might be around 70%. If it is then what meaning does his 70% number have for distinguishing species or kinds? Probably not much.
LikeLike
Thanks for the reply, this was helpful. I’m not all that familiar with how geneticists determine the amount of similarity between two species, but I suspected it wasn’t as easy as comparing every base pair in the genome because of deletions, insertions, repeats etc that occur in otherwise closely related species. Along with the “only 70% similar” figure I’ve heard Creationists throw out that chimpanzees and humans have to be at least 10% similar because the former has ~10% larger genome or that geneticists intentionally ignore the most dissimilar sequences to make chimpanzees and humans look more similar than they are.
LikeLike
With respect to Richard Buggs, I have edited my post. Although he may have leanings toward some version of creation science, my inference about him wasn’t germane to my discussion and so I took that out. I don’t doubt that Buggs is actually a very competent scientists. I know people he has worked with and his publications are quite good. As I said, his values for similarity are likely very accurate given the assumptions he used to make his calculation.
LikeLike
You’re welcome, you may also find Dr. Wile’s post of interest: http://blog.drwile.com/?p=9851. He states for example: “As a result, comparing the genomes of two different species can be very, very tricky.” :)
LikeLike
I didn’t see what you wrote about Richard Buggs but he is known to be a creationist:
http://www.bcseweb.org.uk/index.php/Main/RichardBuggs
LikeLike
Hi Ashley, I did find that page about Buggs after I posted. He definitely has an interesting past. For the purposes of this article that past doesn’t matter much. He is definitely a competent scientists. I think his numbers are accurate in the sense that he isn’t making anything up but as I said in the article the question is whether his method of calculation leads to things sounding more different than they actually are. He needed to use the same methods and calculate other relationships to see if they also are as different to make a meaningful point about genetic distance.
LikeLike
Great article. I agree that YEC’s who try to posit the uniqueness of man are barking up the wrong tree…
LikeLike
Tomkins got his 70% figure because of a bug in the software that he used to do the comparison (“BLAST+”). I re-did his experiment, not excluding any DNA, counting a 100bp indel as 100 differences, and took into account the effects of the bug. I got a final figure of 96.90% +/-0.21%.
My paper is here:
https://www.dropbox.com/sh/dm2lgg0l93sjayv/AAATnWSJdER53EYEYZvcgiwma?dl=0
I submitted it to ARJ almost a year ago now – and Tomkins is the sole peer-reviewer (critiquing a critique of his own work?!?!). The editor doesn’t seem overly keen to push him to respond, and Tomkins has been silent for 8 months now – the ball is in his court. Clearly they are hoping that I just go away and hope that my paper never sees the light of day.
LikeLike
Wow, thanks for sharing this info. I have tried several times to figure out just how Tomkins did this analysis but have always found myself confused. Another genomics friend of mine also took a quick look and although the result didn’t make sense we couldnt’ figure out what is wrong. I have had messages communicated to Tomkins that to bring any meanng to this data he needs to do a apples to apples comparison by doing doing other pairwise comparisons with other species. Even comparing two modern humans with his method I suspected would yield greater differences than he would want to admit. Of course he hasn’t done any further analyses and has responded generally that other genomes are high enough quality to be able to do his analysis. That always sounded like an excuse. Seems that you have found out the real reason why he hasn’t published any other comparisons.
Good for you writing an article. That is a lot of effort but Tomkins and the creationists community in general needs to be willing to review their own claims and have them tested if they want to claim they are doing science. Unfortunately I am not optimistic that Tomkins will respond as I would hope a scientist should respond.
LikeLike
Just finished reading your manuscript. That needs to be published. Fascinating stuff. It answered some questions I have had for a long time about how thee analyses where done.
LikeLike
I agree it needs to be published! Ha! :D
Feel free to hassle Jeff directly – his email address is his first initial and surname at the ICR ORGanisation…
LikeLike