Last week I pointed out that the Bible provides no support for Ken Ham’s contention that massive numbers of species have formed following their departure from Noah’s ark 4500 years ago (YEC Biblical Evolution: I Have A Book That Says Otherwise). Now I’m following up with “observational” evidence from DNA sequences to test whether the patterns of sequence divergence fit the YEC hyper-evolution model.
Let’s review the claim of young earth creationists again. Ken Ham showed the following slide in his debate with Bill Nye. He explained that biblical creation proposes that there were only two of each kind on Noah’s ark and that a kind is best understood as being equivalent to a “family” as understood by taxonomists
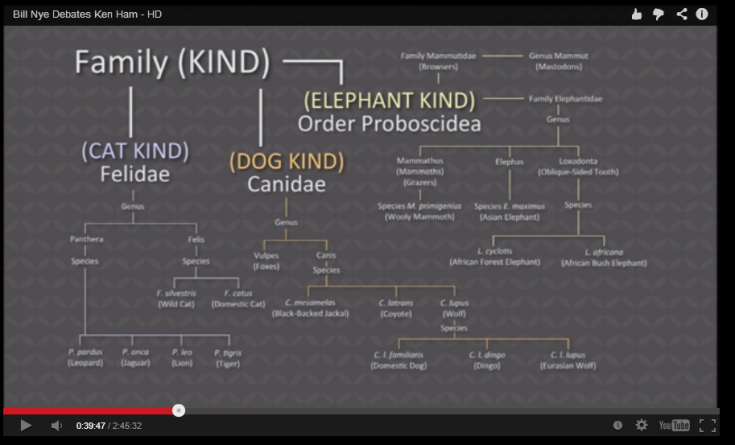
Did all cats, dogs and elephants come from pairs of ancestors just 4500 year ago? Ham and other YECs like to show pictures of domestic dogs and all their apparent variety. Since everyone knows that domestic dogs are varieties of wolves we are supposed to be amazed at how much change can happen within a “kind”. Ken Ham then presupposes that this simple illustration can serve to explain all the variation within canines, felines or any other group of animals. He and other YECs seem to think that the formation of hundreds of new species is just the result of sorting out of genetic variation found in the representatives of each “kind” preserved on the ark.
But just how much variation is there within and between species? I have examined this question before (Of Kinds and Common Ancestors: Comparing Mitochondrial Genomes) but I thought I would do a few more analyses and present the data in figures rather than a table. I have used similar methods as described in that prior article. In brief, I have found a complete mtDNA genome (usually around 16,000 base pairs of DNA) and searched a database (NCBI) to pull out all other known mtDNAs that are similar. Then I have constructed a simply tree showing how similar each mtDNA genome is to one another. I have then scaled the trees so that the branches represent the same amount of difference on all the trees I show below. I am doing this to get at an apples-to-apples comparison of genetic differences between species.

The problem is that Ken Ham doesn’t seem to know much about genetic divergence or how to determine degrees of differences at all. In his talk, he showed a figure of domestic dogs and suggested that these dog varieties demonstrate that large amount of divergence of animals was possible in a short period of time (observational science to Ham) . Well, look at the figure above and let’s see how his observations match up to the genetic reality. I was able to get more than 800 complete mitochondrial genomes representing virtually every breed of dog. Included in these are multiple ancient dog genomes. These are dogs that were sequenced from bones that were hundreds to 10,000 years old. What should jump out at you is how incredibly similar all domesticated dogs are to one another. The mtDNA is a good proxy for their overall genetic divergence and so this is why geneticists would consider domestic dogs highly similar to one another.
The reason domestic dogs look very different morphologically is a matter of changes in a few developmental genes and a couple of genes for coat color, etc. You could say that the differences between dogs are really only skin deep. The vast majority of domestic dog DNA is identical to each other owing to their “recent” evolution from just a few wolves. Conventional evolutionary genetics predicts that domestic dogs would show very little divergence given they have only been separated from wolves for 10 to 20 thousand years. This is not considered enough time (generations) for large numbers of mutations to have built up in dog populations.
Now look at the differences between wolves, coyotes and then the foxes. This is a substantial amount of genetic divergence and this also reflects the millions of differences found in their nuclear genomes. These canines have thousands of different versions of genes and even differ in their number of chromosomes. In other words, domestic dogs may look different from one another but these differences are very superficial. And yet young earth creationists hold domestic dogs up as observational evidence that hyper-evolution can occur within “kinds.”

Now let’s take a look at cats and see how similar they are to one another. For cats species there aren’t as many complete mtDNA genomes, particularly for domestic cats. I am sure that all domestic cats would show no more differences from one another than the domestic dogs show or modern humans (see below). I have scaled this figure so that it is as close to an apples-to-apples comparison with the canines above. You can see that there is quite a bit of genetic divergence between the species of cats. For some perspective, given the amount of divergence observed and dates of fossils it is estimated that lions, tigers and leopards are thought to have split off from the other feline lineages, including domestic cats, some 10 to 15 million years ago.
What we are seeing here is very significant genetic differences among some cat lineages. Within a species like tigers there are differences that reflect different populations which have their own genetic history. Also, look at the hyena sequences. Hyenas have similar skeletal and other morphological characteristics to felines and here you can see that their mtDNA genomes also have similar sequences. However, young earth creationists insist that hyenas are a different kind and thus have no common ancestry with felines. But tigers and house cats do have a common ancestor?

There are only three species of living elephants but there are many complete mitochondrial genome sequences that have been obtained from fossils of mammoths and mastodons. What is really interesting about the mammoths in particular is that we have DNA sequences from mammoths that lived in Siberia, North America and even South America. And these samples are of different ages ranging from 10,000 years to 50,000 years old. The YEC model of origins would certainly predict that mammoths that spread themselves out across the Earth should show differences from one another because they evolved super fast early after the Flood.
What we see is that mammoths mtDNAs are remarkably similar. Now this isn’t surprising in an old Earth model of origins. Despite their being spread out geographically and over time, this is still a short period of time with respect to the life span of a typical species. There is variation among mammoths equivalent to a bit more than seen in people alive on Earth today.
Notice also how different the mastodon sequences are from other elephants. They have a very different tooth structure and other physical adaptions. Yet they lived at the same time and with mammoths in North America. If they really were both derived from the same two common ancestors on Noah’s ark then how did they come to be more different from humans and chimpanzees (see below for comparative data) and yet lived in the same environment? How could natural selection have sorted them into two very different elephants if they had the same resources and lived in the same place?
To give you some perspective on the degrees of mtDNA sequence differences in the prior figures, I did a quick analysis of 500 great ape and human mtDNA genomes. Again, I did this by taking a single human mtDNA genome and having the database (NCBI) pull the 500 most similar sequences and then drawing a tree in which the total number of DNA differences is represented as the branches connecting the individual sequences.

I have scaled the figure so the branches represent roughly the same amount of genetic divergence as seen in the canine, feline and elephant analyses above. From this similarity tree we can make several observations:
1) Modern humans are genetically very similar to one another – we truly are all one race! The genetic divergence of all modern humans (not including Neanderthals) is similar to that among wolves and domestic dogs.
2) Chimpanzees are a very diverse species and quite possibly should be considered at least two species. They may all look the same to you and I but the amount of genetic diversity among chimpanzees greatly eclipses that found among all humans including ancient lineages of humans (Neanderthal and Denisovan).
3) The difference between modern humans and Neanderthals is not very great compared to the total difference between humans and other great apes.
But here is the more provocative result of this simple analysis: Look at how much difference there is between humans and chimpanzees. It looks like a lot but then look above at the canines, feline and elephants, and you will see that the difference between the foxes and wolves, between a house cat and lion and a mastodon and mammoth is greater than the differences between humans and chimpanzees. It seems utterly inconsistent of Ken Ham to tell the world that they have no problem with foxes and domestic dogs having a common ancestor just 4500 years ago when mastodons and elephants, foxes and dogs, cougars and lions exhibit greater genetic divergence than chimpanzees and humans.
In the YEC evolution model, humans started out with eight individuals which would represent greater genetic diversity and potential and yet humans have populated the world in the same amount of time that canines, felines and elephants have had, and yet their total genetic diversity barely exceeds that of domesticated dogs much less the variation observed in these other “kinds.” What is the YEC explanation for why humans have experienced such incredibly low rates of change in this genome? I have no idea.
I think this just highlights the ad hoc nature of their proposals. They propose all canines from a common ancestor not because of any evidence that this has happened very recently but just so they can have only a pair of canines on the ark. But they haven’t thought through the consequences of such a proposal. Genetically it doesn’t make sense and it ends up creating more problems than it solves.
Is there any way you can make those figures darker? They are a little hard to see. Could you clarify what you mean by branch length? Are you referring to the horizontal or vertical branch length? It would also help your case if you could quantify the differences in terms of numbers and/or percentages of nucleotide differences. Is that possible to do with the software you are using?
LikeLike
Yeah, they are difficult to read. I just copied the images right from the NCBI interface where I had them generated. I didn’t have time to really work with them in Illustrator. I mostly just wanted to show the patterns of divergence and that individuals within a species are quite similar. If you go to this post: https://thenaturalhistorian.com/2013/02/26/kinds-baramins-creationism-mtdna-genomes-compared/ you will see that I have quantified the total number of differences between genomes of many different animals.
LikeLike
You probably saw this:
http://www.patheos.com/blogs/exploringourmatrix/2014/02/ken-hams-dog-slide.html
LikeLike
Yeah, I considered using that screenshot as well. I don’t think Ken Ham or anyone of his employees really are able to interpret those research articles. I am sure they realize that the dates are far different but I don’t’ think they realized how the genetic works that leads to the conclusions in the paper.
LikeLike
Hello again. Someone from AiG passed this link on to me, so I decided to reply. I am all for testing the creation model, but it needs to be consistent with its own assumption, not necessarily evolutionary ones.
Could you give me a good reason to believe that mtDNA variation is a good proxy for overall genetic divergence?
Do you realize that you have very effectively shown that mtDNA variation and phenotypic variation are not well correlated? This brings up a serious problem evolutionists have in constructing their phylogenies; very different phylogenies may be suggested depending on which characters are used to construct them. Even different choices in molecular characters can lead to divergent phylogenies. I ran into this a lot when I was researching for the Mammalian Ark Kind article.
Do you realize you are using assumptions of neutral theory and the mitochondrial clock hypothesis to show that the creation model is inconsistent with these assumptions? But YECs know this, so the comparison seems a bit silly to me. One of my articles that touches on this subject is on the AiG website. Rob Carter has written more extensively on this in the Journal of Creation 23(1):40-43 and 70-77, 2009. You will note that we cite from the scientific literature to support our objections to neutral theory in general and the molecular clock hypothesis specifically.
Now you are free to accept neutral theory and the molecular clock hypothesis if you like, but serious engagement with the scientific literature gives me strong reason not to. In addition to what you find in the articles above, the loss of junk DNA and results from population genetics modeling* have stripped evolutionists of a logical, mathematically plausible basis to claim common ancestry between humans and chimps.
http://www.answersingenesis.org/articles/aid/v3/n1/explaining-diversity-created-kinds
Click to access j23_1_40-43.pdf
Click to access j23_1_70-77.pdf
* which includes the reality that genetic drift must be accounted for in an evolutionary model: Rupe, C.L. and J.C. Sanford. 2013. Using numerical simulation to better understand fixation rates, and establishment of a new principle: Haldane’s ratchet. Proceedings of the Seventh International Conference on Creationism. Pittsburgh, PA: Creation Science Fellowship.
LikeLike
Hi jean, you are quite right about mtDNA and its limitations. Having published multiple papers on molecular phylogeny reconstruction including many with mtDNA sequences I very aware of the rate heterogeneity. I have done relative rate tests and other measures of rate heterogeneity between nuclear genes, cpDNA and mtDNA in plants and animals. So I would never claim that all the assumption of neutral theory or mtDNA clocks are fully followed. That said, just because there is rate heterogeneity and not all part of the code are neutral doesn’t mean that sequences can’t be compared at all. YECs need orders of magnitude differences in rates to explain observations. Within orders and genera rate differences are rarely that great. When I talk to my non-majors class about how a gene is turned into a protein I don’t tell them about every single post-transcriptional modification that is made to mRNA before it is translated. Does this mean that what I have told them about translation is wrong? it would if those other things meant that translation was so altered as to not be recognizable for what it does without understanding those complexities. In this case, rate changes within some lineages are relatively minor blips with respect to these comparisons. The general comparisons are rough models that provide general truths.
I understand the objection to molecular clock and I can say that I agree that molecular evolves in a completely clock-like fashion. However, saying there is no perfect clock and providing references against a clock is far from proving that many molecules do exhibit nearly clock-like changes. No one should be using mtDNA for deep phylogeny but it can be used quite reliably within most families and orders which is where my comparisons were. I can use a stopwatch with a seconds hand to time my kids running a 5K with enough accuracy that I don’t care that it is perfect, I can still use it to compare to other kids times and get a pretty good idea of who was faster. But if it were a 40 yard dash I am probably going to want a digital display to 2 decimals. A phylogeneticists has to spend a lot of time picking the right tools. mtDNA isn’t the best in many cases. I only used it as a crude tool and in my foot notes of one of my articles on mtDNA I noted the assumptions involved including molecular clock.
Overall i would say that mtDNA is a reasonable but far from perfect proxy for showing overall genetic divergence among species and within genera. When you get to genera within families it will probably under-represent the overall genetic divergence (I’m talking ration between mtDNA and nuclear as mtDNA is always higher). The mutations will totally saturate the mtDNA molecular at this level or higher.
Not sure how far I can go with this. I think I have read all of Rod Carter’s articles and there are so many things I think he either doesn’t understand or he is not presenting the whole story that I don’t know where to start.
The point of my blog wasn’t that mtDNA is the definitive measuring stick but that it is a crude test that suggests there is something very wrong with creationists models. Just changing the molecular clock rates doesn’t fix the problem. There is far more consistency in the differences than there are surprises and now that we are able to sequence ancient DNA we are able to directly test many of our assumptions and thus far no new radical readjustments to our understanding of how genes change have been forthcoming.
I’m rambling on quite a bit here. I’m just frustrated because I know how complex the topic is and it isn’t just a matter of there is no clock so all comparisons can’t be relied upon. No, comparisons can still be valid and applicable even when we acknowledge there is inherent variability in the system.
Thanks for the feedback. I’m very interested in trying to figure out just what a young earth would mean for genetics but I have thus far found the literature very difficult to understand to this point though not for the lack of trying. But I’m willing to take another crack at it. Joel
LikeLike
Hi Jean, I just realized I wrote that whole reply above thinking that you were replying to a different post. The one I had in mind was this one: https://thenaturalhistorian.com/2013/02/26/kinds-baramins-creationism-mtdna-genomes-compared/ which was where I explained more about mtDNA genomes. There is much more detail here about my simple methods but also my assumptions.
Going back the actual article you were responding too. Yeah, I’m stretching a bit to compare canines with felines BUT I would feel quite comfortable with the variation we are seeing within those two groups as roughly equating to their overall genetic differences (ie. if two speces are twice as different in their mtDNA they are probably about as much different in their nuclear genome – scaled with respect to mtDNA having a higher rate of evolution of course). Just look at those domestic dogs. The impression often given is that they are quite different and they seem to be morphologically but genetically the mtDNAs are nearly all the same and this is true in their nuclear genomes as well. The differences between domestic breeds is tiny and limited to only a few important genes that effect growth and development. When I see that a wolf and coyote are far more different than any two domestic dogs that is reflected in their nuclear genomes as well. Then foxes are WAY more different. For Ken Ham and other to point to domestic dogs as evidence that massive evolution could happen from a few animals departing the ark is disingenuous as domestic dogs are barely different from one another and are hardly evidence of what evolution can or has done to species within a family. He can’t simply extrapolate domestic dogs to all canines without inventing brand new genetics. Does Carter, Tomkins, etc. really want there to be a completely different rate of mtDNA evolution in different canine species? Why would they differ by orders of magnitude necessary to explain canines in 4000 years (remember that many domestic breads have existed at least that long and so they effectively have not evolved their mtDNA while other canines have gone crazy with their mtDNAs. A semi-regular clock over a long time is a far more likely evolutionary scenario. It is on the young earth geneticists to provide an alternative mechanism for why clocks could very wildly among very similar organisms.
LikeLike
Thanks for the response. I don’t have time for a lengthy exchange, but I thought I’d briefly respond to your comment “I’m very interested in trying to figure out just what a young earth would mean for genetics ….”
In this context, it would mean recognizing the difference between data (e.g. measured rates of DNA change between parent and offspring) and model-based inference (including calibrating the rate of molecular clocks based on assuming human-chimp common ancestry). Now clearly, YECs accept within kind common ancestry and so make model-based inferences, but I need to recognize when and where I am doing this.
You do clearly recognize that the YEC model predicts rapid speciation has occurred. If this is true, what would we expect to find in genetics?
1) That random mutation and natural selection are not the major players in genetic changes.
2) That mutation rate (e.g. mtDNA) can change, perhaps by orders of magnitude, under certain circumstances.
In summary, it would mean the the naturalistic assumptions of the Modern Synthesis (neo-Darwinism) are wrong. I certainly have seen rumblings of this in the scientific literature; I expect to see more in the future.
LikeLike
I’m sorely late following up with you. I think there are many tests that can be done to look at expected rates within kinds and hypotheses that can be generated with respect to how rates either varied or would have stayed constant since a Flood bottleneck. In genetics you would likely need both 1 and2 to be true. Now 1 we already know doesn’t explain all genetic changes. Genetic drift can be a potent driver of genetic change over time without natural selection. This is especially true for small populations which I suppose would be true for the initial populations spreading from a central point over the earth. Regarding 2, I am quite certain you would need to hypothesize and find that mtDNA rates can change by at least an order of magnitude.
Probably 2 would have to represent really radical changes in rates of mutation in different lineages. I can think of one example right away. Domestic dogs have hardly any difference in their mtDNA and that includes mummified dogs at least 3500 years old. We also have numerous other dogs from burials that are 4-10 thousand years old. Even if the oldest is really 4000 years old this would mean that the rate of mtDNA change among dogs over 4000 years has been very small. At the same time there are many hundreds of differences among different canine species and wolves from one continent have many times the number of differences even between then than all domestic dogs do combined. So mtDNA apparently is changing extremely rapidly during the same 4000 years for other canines.
Because domestic dogs have been bred apart from one another and in small groups the founder effect/genetic drift should be very very strong in domestic dogs compared to wolves. This suggests and even lower base mutation rate among dogs since any mutations would have a much greater chance of becoming fixed in a dog breed population vs wolves where there is greater gene flow.
I think canines then show that either mutations rates/ fixation rates have been orders of magnitude different among canine lineages or domestic dogs are young (<10,000 or so years) and other canine lineage are old (100s of thousand or millions of years old).
LikeLike