The fact that bacteria are able to develop resistance to antibiotics has been a textbook example of evolutionary processes in action. Mutations and natural selection—two primary agents of change—work together to sculpt new genetic combinations allowing individuals to exploit resources unavailable to bacteria previously unable to resist antibiotic chemicals in their environment. Now you can watch a powerful illustration of E. coli undergoing evolution of antibiotic resistance in a video produced by scientists at the Harvard Medical School. The video below shows a fascinating new way to watch bacterial strains evolve and then examine the mechanisms of the adaptive process in detail.
What have you witnessed? This video is just 2 minutes long but provides a good overview of research just published in the journal Science (see reference below). At a minimum you are seeing the origin of new gene variations—alleles—via mutations and natural selection acting on the phenotypes—the physical manifestations—of those mutations.
The initial colony of bacteria introduced onto the edges of the huge petri plate lacked the ability to resist antibiotics. They were able to survive in the agar at the ends which had no antibiotic but in the next “lane” of agar there was enough antibiotic to kill the bacteria. Any bacteria in the initial population that experienced a genetic change that allowed it to survive in the second lane could free themselves – at least for a while – from the competition for diminishing resources in the first lane and find themselves in a resource rich region with fewer competitors. Hence, evolving the ability to resist the antibiotic would be a favorable change for these bacteria.

Each time a bacteria divides it must copy its genome but in doing so they always make some mistakes—mutations. After a day or two, billions of bacteria have colonized the outer lanes of the large petri dish. Collectively, those bacteria have experienced billions of mutations. Most of those mutations would have been negative (either lethal or reduced their efficiency in some way) or neutral and therefore had little effect on the success of the bacteria. Some may have helped the E. coli compete better for the limited resources in the first lane. Of the billions of mutations only a few occurred in genes that provided some individual bacteria the ability to survive in the presence of the small amount of antibiotic in the second lane. Those bacteria were free to move into the second lane and begin to take advantage of the resources available there. As this new colony grew into the second lane the colonies would have continued to accumulate more mutations. Again, most were negative or neutral but some of those mutations conferred even greater capacity of the bacteria to resist antibiotics allowing those bacteria to move into the third lane.
This same pattern of continued mutations and natural selection continues allowing mutations that provided greater ability to resist antibiotics to accumulate thus allowing additional area with more resources to be invaded.
Mutations are random but natural selection is not.
What is really cool is that not only can you see which lineages of bacteria had mutations that conferred a greater ability to resist antibiotics but because each bacterial lineage can be traced the researches could sample the bacteria in those lineages. They subsequently extracted the genomes of those bacteria and sequenced them to reveal their entire code. By examining of the complete genomes of ancestors and descendants, they located every change (mutation) in them. By characterizing when and where those mutations occurred in the genome they could identify the chronological order of those mutations and therefore recreate the steps the bacteria used to gain resistant to the antibiotic.
You can see from the video that multiple lineages of bacteria from both sides were able to generate adaptations to every increasing antibiotic concentrations but the interesting question is, did they come up with the same solutions to the antibiotic problem? The answer is, no! They found unique ways of solving the same problem. Natural selection is a process that seeks to solve a problem for the organism but the solutions it finds are limited to the available pool of variations from which it can choose. Those variations are usually generated randomly by mutations.
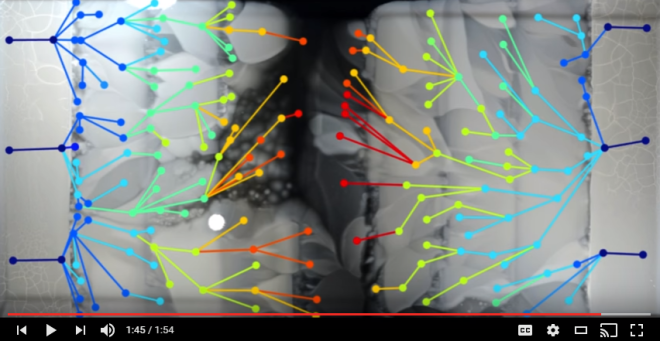
So the mutations were random with respect to locations in the genome but whatever mutations did occur were then sorted by natural selection eventually resulting in new functions. Interestingly, the bacteria that finally conquered the 1000x dose of antibiotic were able to do so not because of a single large change in its genome but as a result of many small steps that added up to high resistance. By introducing the bacteria to a gradually changing environment – from low concentration to higher and higher concentrations – the bacteria were able to take advantage of multiple mutations to make small adaptations along the gradient. The Harvard scientists tested the ability of the bacteria to make the jump directly to high concentrations of antibiotic and that experiment failed to generate resistant bacteria. This showed that the ability to resist high loads of antibiotic required a genetic apparatus that was too complex to originate by a single mutation.
The same thing happens in nature. As long as a population is not thrown into a radically new environment some individuals will have new variations of genes resulting from mutations that will allow them to survive in that new environment after which they can further adjust to the new conditions through additional mutations. However, large-scale changes, such as snakes being introduced onto an island that has many flightless birds who lay their eggs in nests on the ground will result in the extinction of the birds because they are unable to adapt quickly enough to such a dramatic change.
But are we witnessing evolution?
We certainly are witnessing a genetic change in this population of E. coli and thus witnessing biological evolution in action. We are witnessing descent with modification. Young earth creationists will respond that this study is much ado about nothing. This is only demonstrating adaptation and not evolution. After all, this isn’t evolution because the bacteria didn’t change from one kind to another which is what they think must happen to meet their definition of evolution. I expect that they will claim that no new information was created by these changes but rather that the ability to resist antibiotics requires the loss of some other ability and therefore doesn’t constitute real advancement.
The latter would not be an accurate statement though the former is correct: we have not witnessed one species of bacteria becoming something completely different. But this demonstration never claimed to witness the entire process of macroevolution – the origin of new species – but rather allows a powerful witness and test of evolutionary mechanisms that can lead to new species formation. This new research apparatus provided a new way to ask and answer many questions about how adaptation occurs. For example:
Can organisms adapt to new environments? Yes. Nothing new here but the video clearly shows bacteria adapting to an environment in which the ancestral lineages were not able to survive.
Can natural selection act on mutations that improve the fitness of individuals in populations? Yes! Only a couple of the billions and billions of bacteria that experienced mutations had mutations that were useful in that new environment and yet that new environment was able to select those individuals and allow them to colonize that environment.
If the vast majority of mutations are lethal, somewhat deleterious or have no effect doesn’t that mean that mutations are destroying species over time rather than helping to form new ones? No. This is evident in this experiment. From the time that a few bacteria became billions as they grew in the non-antibiotic zone there would have been billions of individual mutations among them. Most of those were likely neutral and therefore had no effect but millions were probably lethal or severely impaired the bacteria. Those bacteria simply did not continue to divide leaving other bacteria with neutral and positive mutations in that environment to perpetuate the colony. So natural selection is sorting out millions of negative mutations in addition to selecting for positive mutations in the new environment. It is these new mutations, however rare they may be, that yield adaptation to new or changing environments that result in real changes to a species over time.
Can mutations produce new interactions among genes? Yes. Direct sequencing of ancestors and descendants which is what this experiment allows and why it is so powerful – beyond the visual aspect – shows that mutations in genes cause them to work together in new ways to produce resistance to the antibiotic.
Do all independent lineages of bacteria that gain antibiotic resistance use the same combinations of mutations to achieve that resistance? No, but they did share some mutations in common demonstrating that natural selection under the same conditions will recognize identical mutations when they occur and select the individuals with those mutations. In other words, natural selection is far from random but rather the arbiter of the value of any mutation.
Do bacteria that become more resistant to antibiotics compensate for that increased ability by losing other abilities? In most cases yes, but in several cases, compensating mutations occurred later in the same lineages that allowed them to gain back all their previous functions making them truly more adapted to the new environment than any of their ancestors.
All of these results demonstrate what was already known about how bacteria may evolve but this experiment helps tease apart the role of mutations and natural selection in new ways. What this experiment so elegantly demonstrates is that new environments cause the sorting of millions of variations in genomes all caused by random mutations when cells copy themselves such that new variations that confer some advantage are preserved in descendant lineages that take advantage of those new environments. This is natural selection in action. This is descent with modification. This is the process of evolution being played out on a small temporal and physical scale.
This new method of observing and testing evolutionary mechanisms has similarities to Dr. Rich Lenski’s work with bacterial evolution. Lenski’s lab has evolved bacteria under the same conditions for over 60,000 generations and documented mutations through that time to show how the bacteria become more fit for their environment and in some cases show that some lineages of bacteria have gained the ability to utilize resources unavailable to the ancestral bacteria. I expect that like Dr. Lenki’s work this new experimental apparatus will provide many new insights as it allows many evolutionary mechanisms to be tested. I am sure that we will be seeing many more videos like the one above in the future.
Reference:

As a person currently under treatment for one of the new “super bugs” and in regular communication with top experts on this topic, I have a few comments:
The above article is incredibly outdated in its assumptions and conclusions. Research in the last 20 years has shown that all of the assumptions of Neo-Darwinist (aka Modern Synthesis) evolutionary thinking are incorrect (!!!!). YES that’s an incredible statement. My wife is well educated in evolutionary population biology (Stanford 1970’s) and was shocked by what has been discovered. Just the discoveries by Barbara McClintock are telling… resulting in a bottom line statement: the genome is an organ of the cell. In fact many now use “gene” in scare quotes because it is all way more dynamic than we have assumed.
A good resource on this: http://onlinelibrary.wiley.com/doi/10.1113/expphysiol.2012.071134/full
Summary of some things that have been found to be false:
(1) “genetic change is random,”
(2) “genetic change is gradual,”
(3) “following genetic change, natural selection leads to particular gene variants (alleles) increasing in frequency within the population,”
(4) “inheritance of acquired characteristics is impossible.”
Consider this statement: ‘it seems that a cell’s enzymes are capable of actively manipulating DNA to do this or that. A genome consists largely of semi-stable genetic elements that may be rearranged or even moved around in the genome thus modifying the information content of DNA.’
This is delightfully upsetting to both YEC and natural selectionists. :)
A friend of mine does advanced cancer treatment research, using transposon techniques. One core, and very practical, principle on which their very effective work is founded (in creating custom treatments that target specific cancer cells etc): there’s almost NO significant new information being developed in the kinds of genetic “mutations” that produce either resistant bacteria or new treatments that attack resistant forms. It’s a very dynamic process of essentially cutting and pasting what already exists, and in particular, discovering sequences in the “junk” DNA that when activated provide helpful or more commonly harmful effects.
Consider some of the more advanced discoveries in recent years, such as Quorum Sensing: bacteria communicate within and between species, and possibly even between kingdoms (?!?). This one is VERY nasty for my current situation. The latest bad-bugs are not detectable in normal lab tests until things are wayyy out of hand. Why? Because the bacteria are completely benign at first. They just replicate, growing their population until a quorum is reached — enough that an attack will more likely succeed (obviously not THAT strategic — we hope — but that’s the effect.) Only then do they create toxins or whatever their attack vector will be. (We’re currently discussing why he felt confident that, while I was initially being treated for an invisible strain, now that another strain showed up he is reasonably sure I don’t also have the invisible strain… a bit scary)
Anyway… given that bacteria under stress can modify their genome up to 100,000 times (per minute? I need to find the paper on this) within a single cell — NOT via descent etc — it is really no surprise that such an encouraging and safe environment such as the above experiment allowed resistance to form.
This is why it is SO important to hit infections HARD and FAST. The bacteria will find a way to resist if they live long enough. Not evolution… defense.
LikeLike
Some truth in some of these statements but the problem is that these generalities don’t address the specifics in this example. Here they sequenced the entire genome and showed specific mutation occurred, where they occurred and their results. There are many other mechanisms for how genomes can adapt but these are not operative here nor have they been shown to be the norm elsewhere but rather the exceptions to the rules. Providing an exception to a rule does not make it the new rule.
LikeLike
Sadly, the article is paywalled. Are you certain they completely sequenced the genome? If so, there should be data on the adv/neut/deleterious proportions, as well as the mutation rates.
What i’m describing is quite operative here. Recent discoveries show that stress — including that encountered in a MEGA-plate scenario — typically induces a pretty radical change in mutations as the organism seeks a way to survive. It’s not at all an increase in reproduction. There are at least two general methods involved: increased mutation (typically via transposition etc.) and decreased error correction. (In reality we’re finding that DNA replication is full of errors but there’s an awesome error-correction/repair mechanism built in. Turn off the error correction and suddenly you’ve got a mutation machine :)
A couple of interesting links (I still have not found my most extreme example) may serve to illustrate the significance of this:
Mutation as a Stress Response and the Regulation of Evolvability
http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3319127/
“Among 787 natural isolates of E. coli from a wide range of habitats worldwide, an astonishing >80% exhibited stress-inducible mutagenesis.”
Inhibition of Mutation and Combating the Evolution of Antibiotic Resistance
http://www.ncbi.nlm.nih.gov/pmc/articles/PMC1088971/
“In this work, we show that preventing induction of the SOS response by interfering with the activity of the protease LexA renders pathogenic Escherichia coli unable to evolve resistance in vivo to ciprofloxacin or rifampicin,”
Bottom line: stress-induced mutagenesis is SO important that they literally stopped the development of resistance by inhibiting the increased mutation rate!
LikeLike
MrPete, are you familiar with Perry Marshall? I haven’t read his book yet but I just learned about quorum sensing from a TED video posted on his cosmicfingerprints blog and a lot of what you are talking about reminds me of the kind of stuff he seems to harp on.
LikeLike
Josh, that’s an amazing coincidence!
I heard about Quorum Sensing from the Infectious Disease specialist who is treating me here… top guy in our region, in continuous contact with CDC. My next appt is tomorrow — if you have questions ask quick :)
Completely independently, I heard about Perry Marshall about a month ago. I’ve seen some of his stuff online, didn’t know he has a book, have had very little time to dig in. I sense that we would agree on many things. AFAIK my focus is different but probably overlapping.
(I’m about to be offline for more than a month for international travel…)
LikeLike
I sure hope that agar plate is destroyed once they have taken their samples.
LikeLike
Minor side note: as an information specialist, I would not accept your statement about information content. “I expect that they will claim that no new information was created by these changes but rather that the ability to resist antibiotics requires the loss of some other ability and therefore doesn’t constitute real advancement.”
Too many assume that the active genome contains 100% of the genome’s information content. In reality, more than 90% of the human genome is non-coded… in my world, commented-out :)
Thus, there’s a huge library of available alternatives ready for use. No new information needed, and nothing needs to be lost by activating some of those sequences.
My cancer-research friend suggests a scary possibility related to this: we are in Devolution. Lots of deleterious mutations are slowly emerging over time… either that reduce our survivability, or that increase the ability of bacteria to attack. Maybe they win in the end after all. After all, much of what we ingest is for our microbiome, not our human cells :)
Who knows… what if God installed a failsafe mechanism in life: go too far off the rails and He allows a meltdown gene to activate…
LikeLike
Joel, you’ve accurately predicted my response…
“that no new information was created by these changes but rather that the ability to resist antibiotics requires the loss of some other ability and therefore doesn’t constitute real advancement.”
You said you agree with the former (no new info) but disagree with the later (no real advancement). Can you shed more light on why you aren’t convinced by the YEC assertion that the destruction of current genetic mechanisms will not yield genetic advancement? In particular, the Science paper found that the most frequently mutated gene on TMP plates was folA, the primary target of TMP. In your opinion, how does the mutational impairment of a folate metabolism gene lead to new genetic innovations? Keep in mind that they found that “Mutations that increased resistance often came with a cost of reduced growth […].” The reduced growth could be compensated for, but only by another mutation (further loss of information). I can’t imagine any scenario where this mutation would lead to speciation, beyond its contribution to geographic isolation. Maybe someone could claim exaptation, but that seems pretty arbitrary here since no new function for the mutated gene was demonstrated.
Thanks
LikeLike
Trevor, I’ll weigh in briefly, then get back to my must-do honey-do’s :)
Your entire paragraph about destruction, advancement, mutational impairment, genetic innovation, growth rates, etc… all has exactly zero relationship to information content.
There’s a precise yet “relative” rather than “absolute” standard definition and metric for “information.” The terminology may sound computer-ish but that’s because most of this work has been done in the computer era. In reality all we are dealing with is some kind of coded representation of facts or sequences of (something). Look up Kolmogorov Complexity or https://en.wikipedia.org/wiki/Algorithmic_information_theory — this is all standard math, even if a bit specialized.
Key to understanding: formal “information content” of X relates to the shortest algorithmic description (eg computer program source code) that can produce X.
This is very satisfying in some cases: adding a second copy of X adds almost no information (if you have two copies of a phone book, what additional info do you have :) )
It’s UNsatisfying in others. Completely random data has more information than a useful message.
LikeLike
Sometimes its good for biologists to standardize their terminology with computationalists. Other times, like this one, the exercise is just distracting. An extra phone book may not be informative to you as an information specialists, but its very relevant to me as a biologist… assuming that the phone book is an allusion to genetic substrate. I’m a functional geneticist, so I’m interested in functional information as defined in the biological sciences. There is no new function reported in the mutant bacteria, only loss of function. So my question remains. How will the loss of genetic function (i.e. mutated folA) result in new function?
I’m asking this with a genuine desire to understand your position, Joel. Thanks.
LikeLike
(I’m Pete not Joel, but we seem to be the only two discussing at this point :) )
My wife is a biologist, and I have enough to be intelligently ignorant :-D, so I think I can be of some help.
Let’s stick with the phone book analogy for a minute, but do understand I am thinking about biology and DNA as I write. “Information” is not the same as “informative” nor “useful” nor “meaningful.” It is simply a sequence of … let’s call them symbols. In a phone book it is character glyphs, plus spaces and line changes and sometimes bolding or other ways to represent… something. For a phone book, we ascribe meaning in terms of names, addresses, and phone numbers.
If I have two copies of a phone book, the only additional information needed to recreated what I have is this: do I need one or two of them? If I can create one, I just repeat once to get another.
Sure, there are lots of other possible results because I have two. I can give one away or burn one or drop it in a lake… and still have one. I can scribble notes in one and still have a pristine copy. I can tear one up and rearrange everything, without affecting the other.
But those are all possible futures. They have no impact on what exists now. What exists now with a second phone book from an information perspective, is that I only have a tiny bit more information: one or two.
Let’s take it one more step. Suppose I add one more bit of info: the second phone book is printed in unreadable IR ink (both on white paper.) The text is all there but it is useless without special lighting.
Again, the information has hardly changed. From a usability and functional perspective, the second phonebook might as well not exist.
One more then back to DNA. We dunk half the phonebook in water, then let it dry. The pages are all there, but are stuck together. Again, essentially no information change but the information is inaccessible and useless.
Back to DNA. And “Junk” DNA. Not transcribed. ~90% of the genome is inactive (you’re the expert, not me. About right?) ALL of that extra DNA is still information. If mutations cause some DNA to stop forming proteins, and other DNA to start… that’s actually a miniscule change in information, even though the organism’s functional aspects may be radically different.
To significantly increase information, requires the addition of significantly new and different coding sequences than what are already there. OR it requires significant degradation of what exists, converting from existing sequences to more randomized sequences. The first is helpful, the second is harmful.
Enough for now. Let’s chew on that much.
LikeLike
Lol, I know you’re Pete. The first part was directed to you; the last sentence was directed to Joel. I want to understand how he thinks about this issue, so I was trying to coax him out. Sorry for the confusion.
LikeLike
(should have written at the end: the first is often helpful, the second is usually harmful)
LikeLike